Receive the latest news, event invites, funding opportunities and more from the Ontario Institute for Cancer Research.
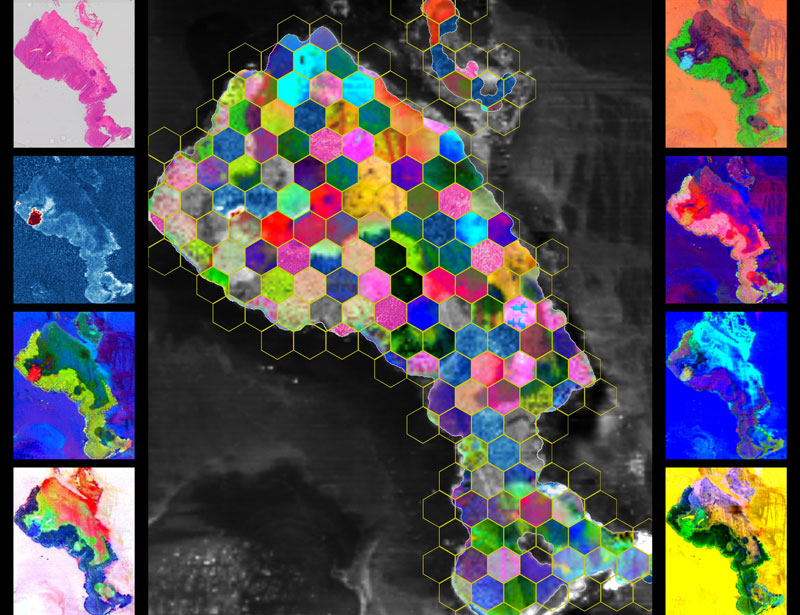
Molecular Mosaic
by Amoon Jamzad, Queen’s University
We analyzed a slide of human colorectal tissue using DESI mass spectrometry imaging, a method that measures thousands of molecules directly on the tissue—no dyes or labels. The result is a set of spatial biochemical maps showing where molecules concentrate in and around cancer. With MassVision, code-free platform, we created complementary visualizations: known molecules (targeted), data-driven discoveries (untargeted), local signal differences (local contrast), and maps aligned with a histopathology (co-localized). We then arranged mosaics of these maps that preserves the tissue’s true layout. All images are real measurements, not simulations. Making these molecular patterns visible can help researchers see cancer and tissue heterogeneity and generate ideas for diagnosis and treatment.
Artist statement
A slide of tissue, a chorus of signals: cancer’s chemistry becomes tiles and tones, each molecular note adding color to a quiet, defiant song.
Tools and techniques used
desorption electrospray ionization mass spectrometry (DESI-MSI) for imaging, MassVision for visualization of ion signatures, Python for mosaic extraction/arrangement
Credits
Amoon Jamzad, Postdoctoral fellow, Med-i lab, School of Computing, Queen’s University, Kingston, Ontario

InterConnections is supported by Illumina