Receive the latest news, event invites, funding opportunities and more from the Ontario Institute for Cancer Research.
The Diagnostic Development Program looks for new and more personalized ways to diagnose and treat cancer. We work with doctors and scientists across Ontario and around the world to improve people’s experiences with cancer.
When it comes to diagnosing and treating cancer, one size does not fit all. The current treatment options for many cancers only work for a limited number of patients, and it is sometimes unclear who will benefit from a certain treatment and who won’t. We are trying to understand why these differences exist so we can create more personalized cancer care that is tailored to a specific person and the biology of their tumour.
We use an approach called “omics”, which looks at many different molecular features of a person. For example, genomics looks at a person’s DNA and RNA. Proteomics looks at the proteins a person produces from their DNA and RNA templates. Data from each of these, when compared in larger studies, can help find the best course of treatment for a person. We look for clues in this data, known as ‘markers’, that can add to and improve decisions about a cancer patient’s treatment . We are also looking at new ways to collect omics data from samples of blood, urine and tumours, which are less invasive than taking tissue samples.
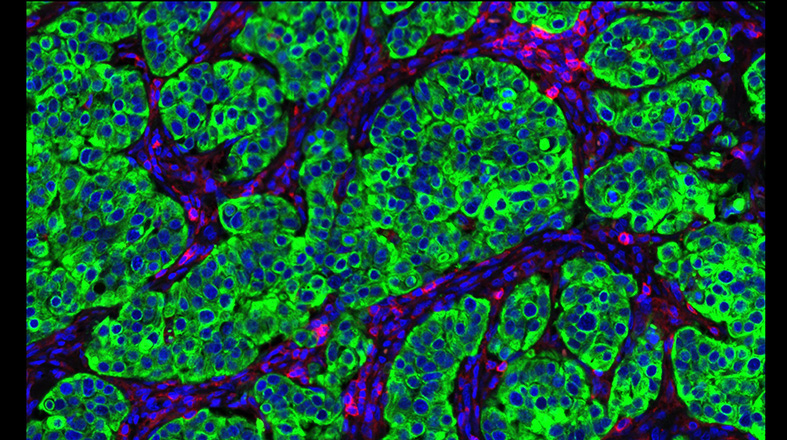
The Diagnostic Development Program drives the implementation of precision medicine through biomarker discovery and validation and the development of assays to guide personalized diagnosis and treatment. A key component of our mission is to extend support and expertise to the cancer research community through active collaborations across Ontario and beyond.
The program’s goals are to :
Develop novel diagnostic assays
Develop new technologies to accelerate diagnostic development in partnership with local researchers and diagnosticians
Partner with OICR and Ontario researchers to enhance diagnostic assay development
OICR is committed to strengthening cancer research in Ontario through collaboration and broadening access to our technology infrastructure, expertise and resources. Please visit OICR’s Collaborative Research Resources directory for more opportunities to collaborate.
Collaborations on tissue-based research can benefit from the technology platforms currently available through the Diagnostic Development Program.
The Diagnostic Development Program operates in a dedicated laboratory suite for tissue handling, molecular biology, histopathology, next generation sequencing, and spatial proteomics with state-of-the-art instruments in support of the following key initiatives:
The Diagnostic Development Program is focused on the development and validation of objective clinical assays to improve therapeutic selection for early cancers. Alongside the program’s core deliverables, we support or have supported research in breast, prostate, ovarian, bladder, pancreatic, endometrial and brain cancers. We focus on tissue-based technologies using human samples in a variety of formats: from formalin-fixed, paraffin embedded (FFPE), fresh-frozen (FF) and liquid tissues (bloods and urine).
We offer a range of services that cover/encompass from the start of tissue handling, derivatization, histology, genomic, transcriptomic, proteomic and downstream analysis, assisting with all aspects of a project.
With human tissues at the core of most clinical cancer research, we offer a breadth of fee-based services helping researchers to get the most out of their material through our team known as Tissue Portal. We offer the following services for FFPE, fresh frozen and liquid biopsy tissues:
Nucleic acid extractions (DNA and RNA)
Histology
Quality control and assurance
Correlative sciences biobanking
Our protocols are compatible with next generation sequencing, immunohistochemistry, NanoString, DSP and other downstream assays.
OICR enables research by providing expertise, advice, and access to research services on a cost-recovery basis. In the focused areas of biomarker discovery and clinical translation, the Diagnostic Development Program has ongoing research projects and associated technologies in the areas of:
Genomic profiling
Transcriptomic profiling
Spatial Proteomics

Contact diagnostic.development@oicr.on.ca to see how we can help you with your project.
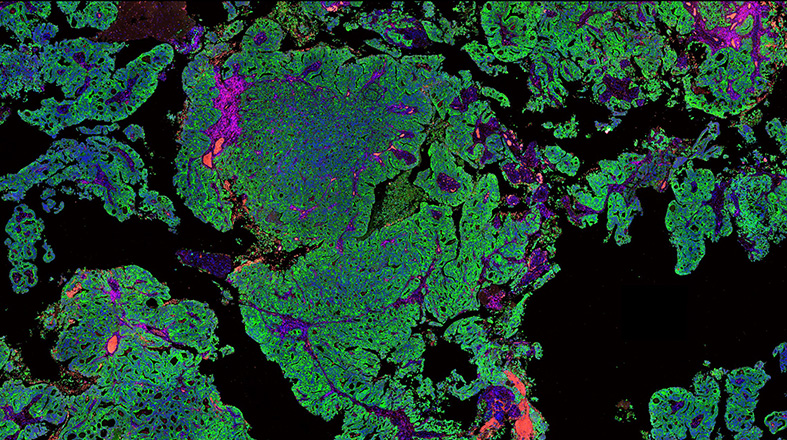
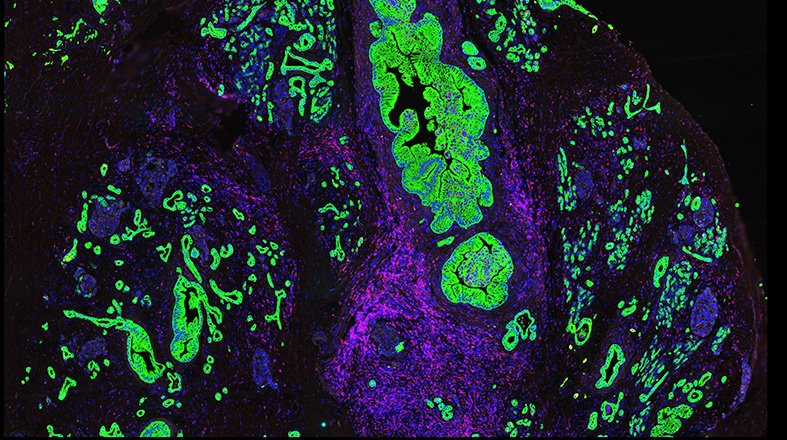
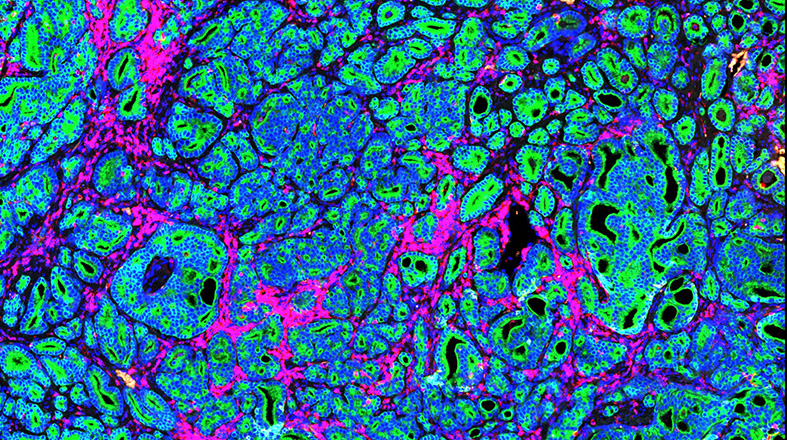

Window-of-Opportunity clinical trials – Learn more about this program
Radiomics – Learn more about this project
UrinePRONTO – Learn more about this project













